
Suppose you need to compare two versions of a large complex chip layout. The second version has been processed in one way or another (say sizing, or OPC) and you want to make sure that the geometric manipulations did not result in any dropped polygons or other unwanted artifacts.
You could run a very exact Boolean comparison but find that such a comparison runs for hours on your largest machine.
Is there a faster way?
The answer is Yes — assuming you don't need the accuracy that a Boolean comparison can provide.
Suppose your sizing or snapping corrections were on the order of 0.1 um and you don't want to see any differences based on that operation. What you do want to see is if a polygon on the order of 2 um or larger was dropped or clipped.In that case one could rasterize both files at 0.5 um. In that case, differences of 0.1 um are "invisible" to the rasterizer but differences of 2 um will result in visible errors.
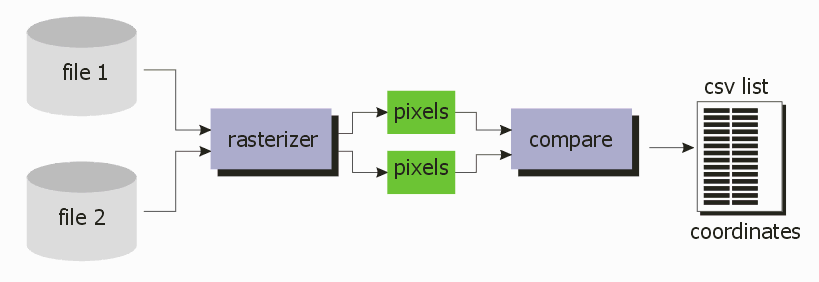
Rasterizing a complex layout at 0.5 um could still take a lot of time but the QisMLib/QisMRaster libraries are heavily multi-threaded and assuming one has a powerful workstation — you do, don't you? — the same comparison that could take several hours using a Boolean approach can take 10 minutes using bitmap comparison.
Artwork created an application,QisMRastCOMP2Files, that does exactly this and which relies on our QisMLib library with the QisMRast extension and a new bitmap comparison library.
Here is a finer grain explanation of how this works.
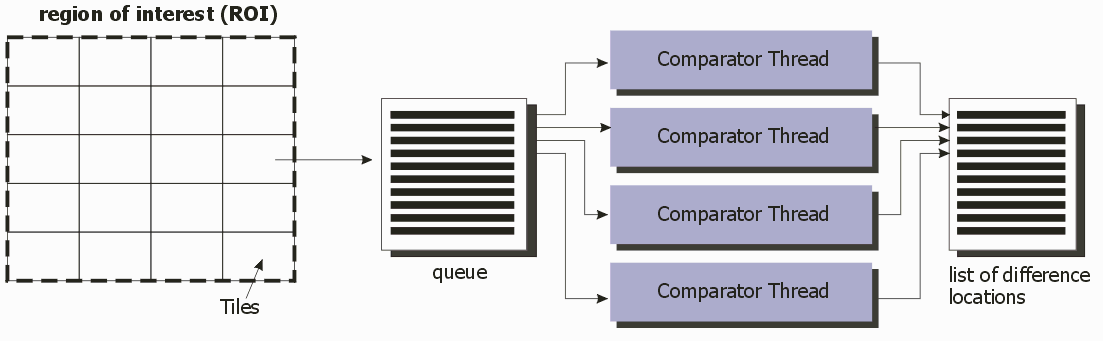
Each Comparator thread handles the data specified by an tile in the queue. When the comparator is complete, it returns to the queue and gets the next available tile. Eventually all the tiles are processed and the results are made available.
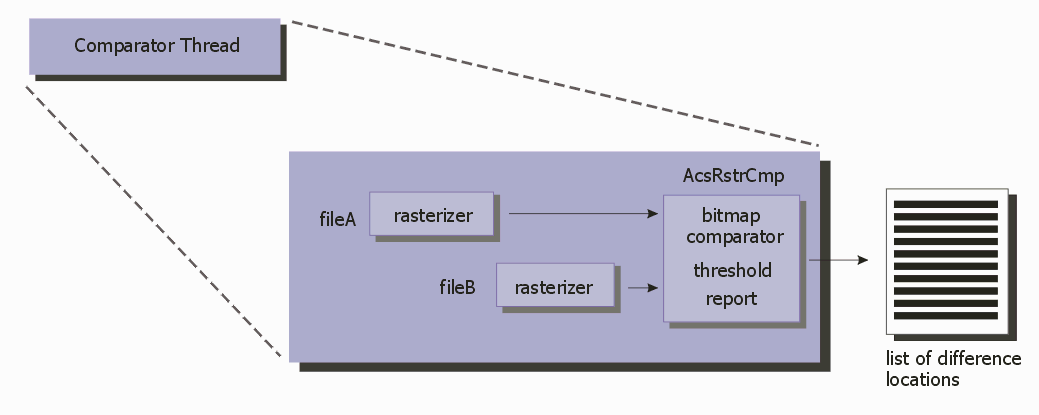
Notes
qismrstrcmp2files.exe +inputA:fileA +inputB:fileB +out:{output-base-path}whereArgument | Description |
|---|---|
+inputA:fileA |
can be either a GDSII file, an OASIS file, or a dbload memory map |
+inputB:fileB |
can be either a GDSII file, an OASIS file, or a dbload memory map |
+out:{output-base-path} |
a path to the directory where the results should be written. |
[-cellA:{first-file-cell}] |
View cell for file-A. If not specified, the deepest top cell is used. |
[-cellB:{second-file-cell}] |
View cell for file-B. If not specified, the deepest top cell is used. |
[-layers:{layers-to-compare}] |
Comma-separated list of layer(s) or layer:datatype(s) to compare |
[-resolution:DPI,{dots-per-inch}]
|
Specify the resolution for generating raster images |
[-tile:NXY,{nx},{ny}]
|
NXY |
[-limit:{limit-difference-counts}] |
Terminate comparison as soon as at least {count} differences are found |
[-sliver:{sliver-value-px}] |
Ignore differences smaller than {value} pixels |
[-ntw:{n-exploders}] |
Specify the number of window/processor threads. |
[-ntp:{n-raster-threads}] |
Specify the number of threads per window for raster operations |
[-roi:{llx},{lly},{urx},{ury}]
|
Only compare a specific region from both files |
[-csv] |
Generate a .csv file which contains lines (cellname,layer:datatype,x,y) for each difference |
[-gds:{datatype-offset}] |
Generate a GDSII file with a text marker at locations where differences were found and polygons from both files in that vicinity. |
[-log:${NEW_LOG_FILE}] |
Enable logging and specify the log file path |
[-silent] |
Do not write any messages to stdout/stderr |
[-verbose] |
Produces extra information in the log file. |